pGreen Bacterial Transformation Prac
AUSTRALIAN CURRICULUM ALIGNMENT:
-
The use of recombinant plasmids as vectors to transform bacterial cells.
BACKGROUND:
DNA can mutate spontaneously or after an error is made in DNA replication. Biotechnologists have developed methods of controlled DNA mutation; such as, intentionally mutating cell DNA to alter how the cell behaves. However, it is also possible to transfer DNA from one organism into another. This method, called genetic transformation, uses an engineered molecule of DNA to transfer a gene or genes from one organism to another so that the organism is capable of producing the protein encoded by the transformed gene. One application of this process is the transformation of the human insulin gene into bacteria which enabled large scale production of the insulin protein. Genetic transformation in higher plants and animals is a costly and complicated process; however genetic transformation in Escherichia coli (E. coli) bacteria is much simpler and a great practical for students.This practical involves transforming bacteria (E. coli) with a gene from Aequorea victoria; a bioluminescent jellyfish. If successful, the green fluorescent protein (GFP) of the Aequorea victoria will be expressed in the bacteria; causing them to glow bright green under UV light. Using bacterial plasmid-based genetic transformation, students will acquire the tools to transform E. coli bacteria to express new genetic information employing a plasmid system and applying mathematical routines to determine transformation efficiency. By playing an active role in the process of manipulation of genetic information, students will gain a greater understanding of how DNA operates; allowing them to consolidate previous learning such as cell structures of bacteria; structure and function of cell membranes, enzymes, and DNA and RNA; transcription and translation.
PREPARATION- LAB TECHNICIAN
- (If you have the kit BT1.00, skip to step 4) To prepare 1L of 50mM calcium chloride solution, check the molecular weight (MW) of your CaCl2, and multiply by 0.05 to work out the mass needed. Add the appropriate quantity to 1L of sterile water. Stir to dissolve and refrigerate until ready to use.
- To make Luria broth, dissolve 2g of LB powder in 100 mL of distilled water. Autoclave at 121°C for 15 minutes to sterilise.
- If pouring your own LB/Amp plates, add 60 mg of ampicillin powder to 8 mL of sterile water and shake well to mix. Add the ampicillin solution to 400 mL of warm LB agar solution and shake to mix before pouring the plates.
- Prepare two LB/Amp agar plates, two LB agar plates, and one E.coli starter plate per student workstation. This must be done no more than 12-20 hours before the student practical. We recommend preparing the starter plates on the afternoon before the student activity.
- To make E.coli starter plates, flame a sterile inoculation loop and aseptically transfer E.coli from the broth to eight LB agar plates. Follow the technique required to generate single colonies.
- Thaw the plasmid just prior to the class and centrifuge for 5 seconds to collect all liquid at the bottom of the tube.
- Prepare a water bath at 42°C if you have one, or set one up using beakers with hotplates and thermometers.
METHOD-STUDENT ACTIVITY
Note: To use aseptic technique, wipe your bench down with ethanol (or bleach), and keep your work near the
Bunsen burner to waft potential contaminants away from your materials.Preparing the transformation solution
- Label the transformation tubes ‘+’ (+ plasmid) and ‘−’ (− plasmid). Keep the tubes cold by placing them upright in the ice bath. Keep the tubes capped at all times except when in use.
- Add 250μL of ice cold CaCl2 solution to each transformation tube, using a sterile transfer pipette. Maintain the temperature by placing the tubes back into the ice bath.
- Use a sterile inoculation loop to transfer a single colony of E. coli from the starter plate to the ice cold CaCl2 solution in the ‘+ plasmid’ transformation tube. To dislodge the E. coli cells from the loop, spin the loop rapidly in the solution. Check whether your E.coli has transferred successfully – it should be visible in your tube.
- Suspend the E. coli in the CaCl2 solution by drawing the solution in and out of a pipette by squeezing and releasing the bulb several times. Do not incorporate air bubbles in the liquid or allow any liquid to splash up the sides of the tube. The solution should start to become milky white as the cell mass is suspended. To check that there are no lumps or particles in the tube, hold it up to the light; then return the tube to the ice.
- Repeat steps 1 and 2 to transfer a single colony of E. coli from the starter plate to the ice cold CaCl2 solution in the ‘– Plasmid’ transformation tube.
- Your teacher or lab technician will bring the plasmid to your workstation. Using a micropipette, transfer 10–μL (0.01–mL) of plasmid solution to the ‘+ plasmid’ transformation tube. Add the plasmid directly to the liquid in the tube without allowing it to touch the sides.
- Immediately return the tube to the ice bath and mix the plasmid into the bacterial suspension by placing a sterile inoculation loop into the liquid and rapidly spinning it. Incubate the two tubes on ice for 15 minutes.
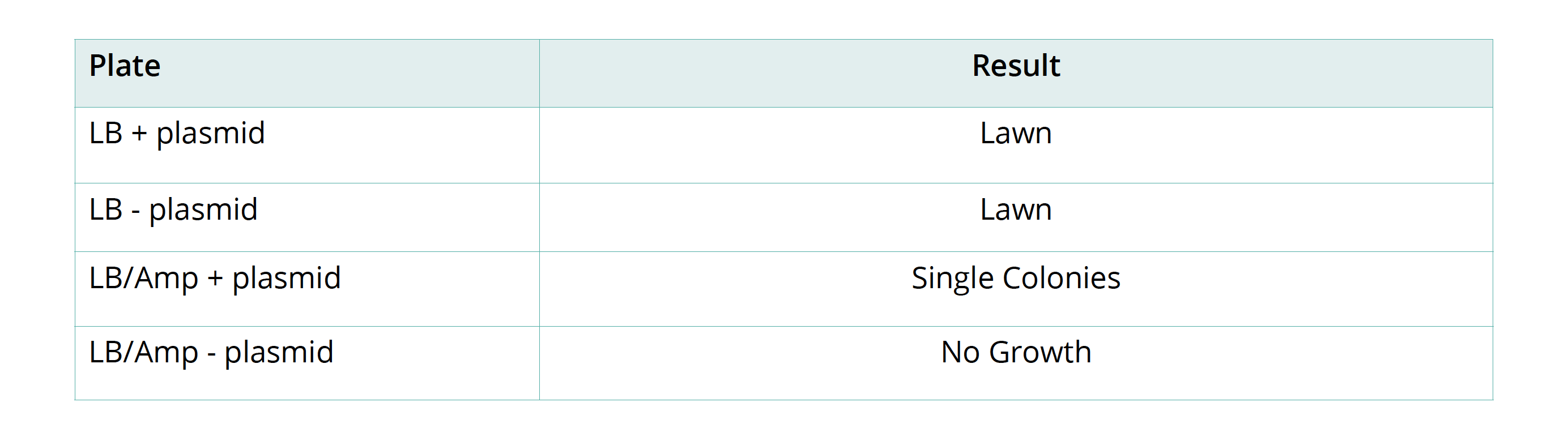
- Label the two agar plates containing Luria broth ‘LB + plasmid’ and ‘LB − plasmid’. Label the two agar plates containing Luria broth with ampicillin ‘LB/amp + plasmid’ and ‘LB/amp − plasmid’.
- After 15 minutes of incubation, remove the two tubes from the ice bath and transfer both of them to a warm water bath (42°C). Hold them in the bath for 90 seconds and make sure the tube caps do not become fully submerged in the water. Gently agitate the tubes while they are warming up in the water.
- Immediately transfer the tubes to the ice bath when the time is up. Allow the tubes to rest in the ice bath for at least 1 minute before continuing.
- Using a sterile plastic pipette, transfer 250 μL (0.25 mL) of Luria broth to each tube. Mix the liquids at the base of each tube by gently grasping the top and tapping the base with your finger.
- Allow the tubes to recover for 10 minutes in a microtube rack at room temperature.
- Use a sterile plastic pipette to transfer two drops of liquid from the ‘+ plasmid’ tube to the ‘LB + plasmid’ plate. Use a sterile spreader to quickly spread the liquid evenly over the plate.
- Use a sterile plastic pipette to transfer two drops of liquid from the ‘+ plasmid’ tube to the ‘LB/amp + plasmid’ plate. Use a sterile spreader to quickly spread the liquid evenly over the plate.
- Use a sterile plastic pipette to transfer two drops of liquid from the ‘− plasmid’ tube to the ‘LB − plasmid’ plate. Use a sterile spreader to quickly spread the liquid evenly over the plate.
- Use a sterile plastic pipette to transfer two drops of liquid from the ‘− plasmid’ tube to the ‘LB/amp − plasmid’ plate. Use a sterile spreader to quickly spread the liquid evenly over the plate.
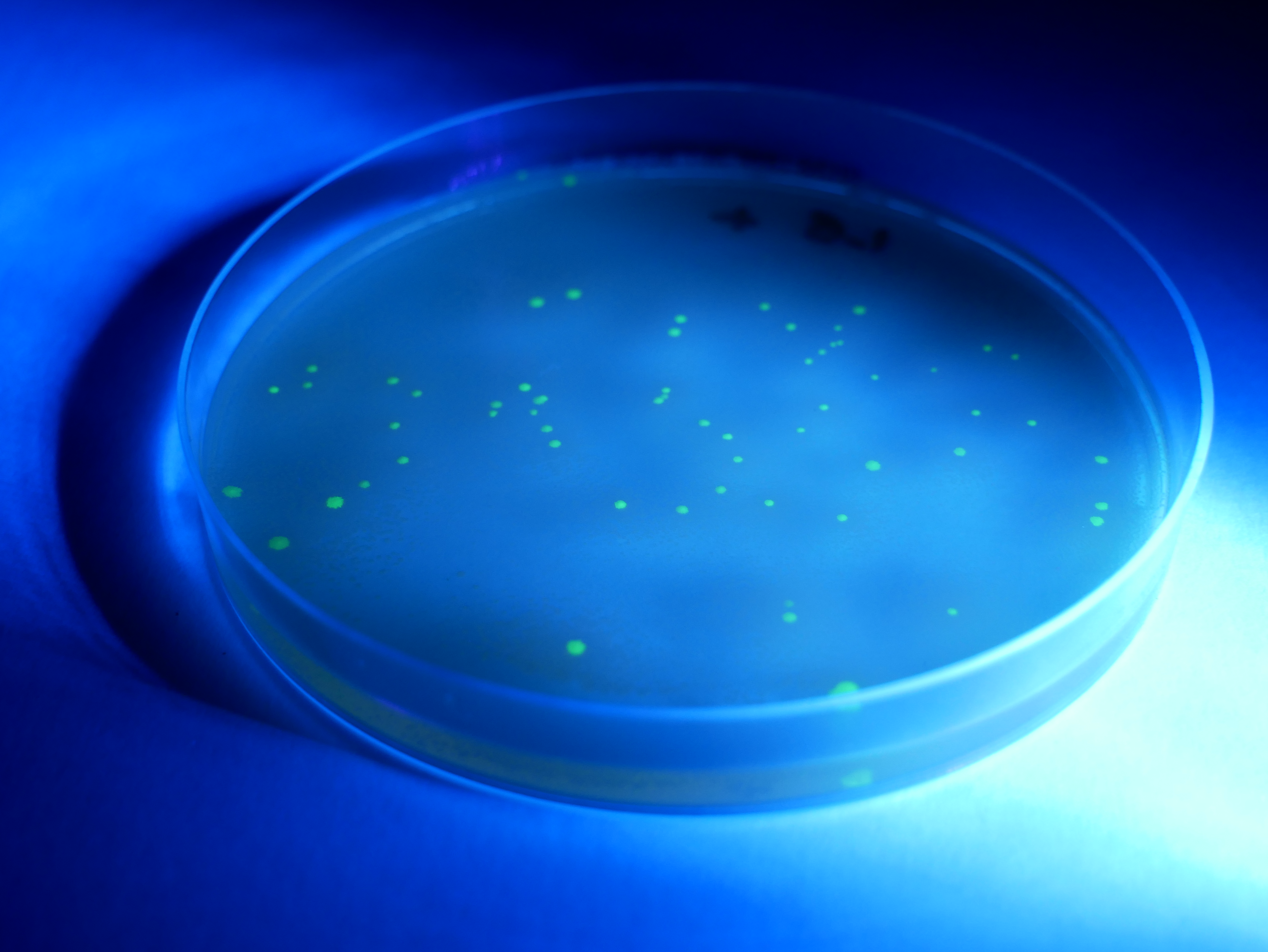
- Secure the lid of each Petri dish to its base with tape. Leave the plates to rest on the bench for 5 minutes and then place them in a 33°C incubator for 24–36 hours. Inspect the growth after this time. You should see either a bacterial lawn, single colonies, or no growth on the individual plates. Take the plates into a dark room to observe evidence of fluorescence in the transformed colonies. Use of a UV light may enhance the fluorescence.
- To count the number of individual colonies, mark the lid of the Petri dish above the colony with a marker as you count it. If cell growth is too dense to count individual colonies, mark that plate as a lawn. Record your results in the results table.
OBSERVATION AND RESULTS
Remove the plates from the incubator to check the results.
INVESTIGATION
-
Why is the plasmid solution placed on ice for 5 minutes?
- Which plate is the control in this experiment? Explain your answer.
- Explain the function of the Luria broth.
- What is the purpose of incubating the cells at room temperature?
- Explain what a plasmid is.
- Explain how the DNA plasmid is put in the bacteria. What is the advantage of being able to do this? (Consider what the plasmid DNA allows the bacteria to do.)
- Explain how we are able to identify that the plasmid DNA is in the bacteria.
EXTENSION EXERCISES
- Write a conclusion of your investigation, including a short discussion of your results.
- Investigate how genetic engineering and bacterial transformation enables the advancement of medical treatments for example in insulin creation.
TEACHER TIP
You may like to use a UV torch to enhance the fluorescence. This will make the colonies easier to count.
 Time Requirements
Time Requirements
- 90 mins
 Material List
Material List
- pGreen Bacterial Transformation Kit
- and
-
Ice bath
- Fine point marker pen
- Micropipette, 10μL (e.g. G40.681)
- Tips for 10μL micropipette (e.g. G40.62)
- Waterbath at 42°C
- Stopwatch
- Test tube rack
- Adhesive tape (to seal plates)
- Microbiological waste disposal bag (e.g. E5.37)
- Thermometer
- Micro centrifuge
- Bunsen burner
- Wire inoculation loop (e.g. E3.10)
- Incubator (e.g. E7.30)
- Boiling water bath (to melt LB agar)
- Microtube rack to hold plasmid (e.g.G40.95, G40.97)
 Safety Requirements
Safety Requirements
-
Some bacteria may cause disease, so assume them to be pathogenic. Note: E. coli MM294 is a commonly used
laboratory strain of E. coli and is safe to use in schools. Wear appropriate personal protective equipment at all times, including eye protection and gloves. Wash your hands thoroughly at the end of the investigation. Decontaminate benches before and after the investigation. Flood any spills with bleach. - Micro-organisms will grow on the agar plates. Do not open agar plates once they are securely taped. Dispose of agar plates appropriately after autoclaving.
![]() Reference Kits
Reference Kits
